I mentioned in an earlier post that sections of our genome have no genes. After the Human Genome Project finished in April 2003, we were left with a list of about 20,000 genes [1]. But that only accounted for 2% of the genome. That leaves 98% that’s non-coding – 98% of our DNA that’s not a gene – seemingly without a function [2].
This result was confusing and a little disappointing. For years, scientists assumed these sections were worthless noise – ‘junk DNA’, but the junk was passed down through generations. Why would life bother to keep worthless DNA? Even more confusing, complex organisms have more junk DNA. Humans have 98%, Flies have 83%, while bacteria have only 12% [3]. If it’s useless, why do we have so much of it?
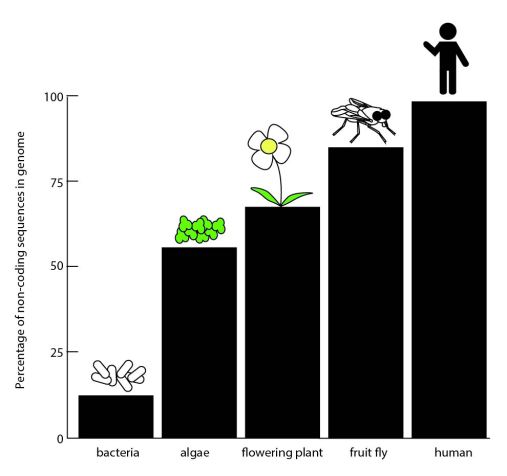
As complexity of an organism increases, the percentage of non-coding sequences within their genome increases [3].
A project called ENCODE, Encyclopedia of DNA Elements, tried to decipher these non-coding sequences by cataloging every bit of the genome. They found that, instead of 2%, closer to 80% of the genome did something. But instead of encoding genes, non-coding DNA regulated each gene [4]. There are roughly 3 million sites in non-coding regions where proteins can stick to DNA. Each of these sites are like switches – proteins bind to a site to turn a gene on or off.
Some of these proteins are called transcription factors – PAX6 is one of them. It’s part of a family of Pax genes that are master regulators of important structures like the eyes [5]. PAX6 directly binds to sections of DNA right before a gene (called a promoter sequence). Certain genes can only be expressed if PAX6 binds, just like an on/off switch.
It’s actually a little more complicated. To turn on any gene, a minimum of 6 proteins need to attach to the promoter sequence. Minimum of 6, but more can be added to increase or modify expression. With up to 20 unique protein binding sites in the surrounding non-coding regions, each gene is controlled by its own complicated locking mechanism [6].
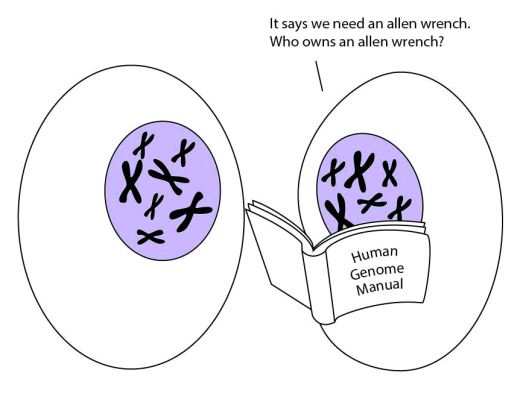
When we first sequenced the human genome, many people thought it would read like a manual. These non-coding sequences confused things – it seemed like the manual was filled with nonsense. But these non-coding sequences are closer to an actual manual than the genes themselves. The genes are the parts, but the non-coding sequences are the instructions on how to put those parts together.
References
[1] “The Human Genome Project Completion: Frequently Asked Questions””. NIH. 30 October 2010.
[2] “Junk DNA – Not So Useless After All”. Time. 6 September 2012.
[3] Mattick, J. S. (2007). “A new paradigm for developmental biology”. J Exp Biol 210 (9): 1526-47. PMID 17449818
[4] “Getting to Know the Genome”. The Scientist. 5 September 2012.
[5] Fernald, R. D. (2004). “Eyes: variety, development and evolution”. Brain Behav. Evol. 64 (3): 141–7. PMID 15353906
[6] “Mysterious Non-Coding DNA – ‘Junk’ or Genetic Power Player?”. PBS. 7 November 2011.
[…] Every gene is made up of several sections of coding DNA (called exons) separated by sections of non-coding DNA. Only the exons are used to build a protein. Of the 12 exons in the GLO gene, humans have lost 7 […]
LikeLike
[…] The bands on each chromosome are made by staining with Giemsa stain [2]. Chromosomal regions rich with adenine and thymine base pairs (AT-rich) stain darker in Giemsa stain. Regions rich with guanine and cytosine (GC-rich) stain lighter. However, most genes are found in these lighter-stained, GC-rich regions – very few genes are found in the darker regions. Which begs the question: if there are almost no genes in these darker regions, what’s that DNA for? (More on that in a later post.) […]
LikeLike